How to Breach Genetic Privacy
Massive parallel sequencing technology has opened up endless possibilities in areas such as diagnosing clinical conditions, finding new drug targets, predicting disease risk and fighting crime. A room with twenty modern sequencing machines can sequence around a thousand human genomes per day. Most practical applications require knowledge of only a tiny section of the genome, which means that the rate at which genetic information can be acquired is truly astonishing. With it comes serious ethical considerations. What happens if your genetic information leaks and can be accessed by employers, insurance companies or adversaries with an axe to grind?
Erlich and Narayanan (2014) describe some of the techniques that can be used to breach the genetic privacy of individuals (with real-world examples of exploits) and discuss some of the methods that can be used to safeguard it from intruders.
How adversaries can breach genetic privacy
There are three larger categories of attacks: based on identity tracing, attribute disclosure using DNA, and completion attacks. Identity tracing is based on meta-data from scientific research, such as genotypic sex, date of birth, zip code and surname. Attribute disclosure attacks are based on accessing the genetic information of a person and then matching it against an anonymous sample linked to sensitive information. Finally, completion attacks allows the inference of target genotypic information based on other areas of the target genome or the genomes of relatives.
Identity tracing attacks
Identity tracing attacks starts with genomic information from an unknown individual. However, this is usually associated with metadata in the form of quasi-identifiers, such as genotypic sex, age, date of birth, zip code, surname and so on. Armed with this information, the adversary can drastically narrow down the range of possible targets to a small group, and then pin-point the individual with the help of information found social media websites such as Facebook. This is done with a wide range of techniques, such as surname inference, DNA phenotypic, demographic identifiers, pedigree structure and side-channel leaks.
Surname inference: because of cultural traditions, the surname is often passed patrilineally, which therefore cause a correlation between surnames and Y-chromosome haplotypes. If an attacker has an unknown sample, but can get an imperfect match in a public genealogical database, that attacker will now know the surname of the target with high probability and that the target is a close relative. With the help of social media like Facebook, the attacker can reduce the number of possible targets to a handful. This technique has been used to expose the identity of over 50 people who took part in the 1000 genome project and all it required were five surname inferences.
DNA phenotyping: some phenotypic features, such as eye color and height, can be predicted from DNA samples from unknown individuals. However, genetic variation currently explain only a fraction of the heritability of these traits, and there are few to no large databases that contain e. g. eye color data or height for a sizable proportion of individuals. Even files from programs such as Microsoft Word, will contain identifying information.
Demographic identifiers: demographic information are highly effective at narrowing down the search space of possible targets. If an adversary has genotypic sex, date of birth and zip code, he can identify over 60% of individuals in the United States. The most dangerous demographic quasi-identifiers, based on how much they narrow the range of possible targets, are zip code, surname and date of birth.
Pedigree structure: about 30% of pedigree structures in one study were unique and this means that pedigrees contain a huge wealth of data that can be used for malicious purposes. Pedigree structures become even more powerful when combined with dangerous quasi-identifiers.
Side-channel leaks: sometimes, the way that genetic data are organized in the databases can leak quasi-identifiers. For instance, the files that contained genetic information of participants in the Personal Genome Project had default files names that contained the first and last name of the participants and these were uploaded by the users to their public profiles. Accession numbers can also reveal disease status of patients or geographical locations if they are predictably assigned.
Attribute disclosure attacks using DNA (ADAD)
ADAD are based on matching the genetic information from a person with an anonymous sample linked to sensitive information, such as drug abuse or medical conditions. This works both on actual genetic data, summary statistics like allele frequencies, effect sizes and information about linkage equilibrium/disequilibrium as well as gene expression data.
n = 1: imagine going to a job interview or a meeting with an insurer. They, unbeknownst to you, collect your DNA from saliva left over on a cup and so now they have your genetic information. They sequence it and carry out searches against large databases of medical conditions or medical history and they find a hit. Apparently, you have a pre-existing genetic condition, or got arrested for smoking cannabis when you were a youngster. You don’t get job or the insurance because of this, but they give you some nonsense rationalization for rejecting your application. The attractiveness of this idea comes from the fact that only a couple of hundred SNPs are required to find any individual.
Summary statistics: even statistics such as allele frequencies, effect sizes and linkage equilibrium or disequilibrium can be used for ADAD. For instance, the allele frequencies will bias any group that the target is in, and this is especially hazardous for rare variants.
Gene expression: this method infers the allelic distribution from gene expression data and connects this with the target and this works essentially like a complicated version of n = 1.
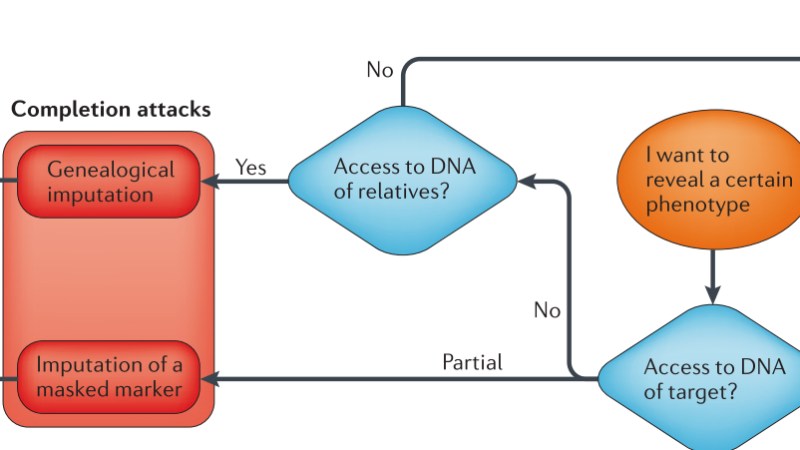
Completion attacks
Completion attacks can use available information to expose hidden or unavailable information. Imputation of masked marker reveals the nature of genes that are hidden by the nature of sequences close to it. Genealogical imputation infers genetic information for a target based on relatives and social media even if the intruder does not have any knowledge of target DNA.
Imputation of masked marker: James Watson, the co-discoverer of the structure of DNA, published his genome on the Internet a few years back. He decided to masked his APOE gene to avoid people knowing about his risks for late-onset Alzheimer’s disease. However, because of linkage disequilibrium between different genetic markers it is possible to unveil the masked region from reference data sets where complete genetic information is available. So if you find a marker in the region outside the masked region and you know that is tightly associated with a particular marker in the masked region, you now know the nature of the masked region with high confidence. Watson’s genome has subjected to such a completion attack as a proof-of-principle by researchers, but genomic regions around Watson’s APOE gene was removed from the published genome as a result. In the future, as more genetic correlations are discovered across the genome, considerably larger segments will have to be masked and probably not just in the close vicinity of the gene.
Genealogical imputation of a single relative: in this exploit, the adversary does not have access to the target DNA, but only to the DNA of a single relative (as determined by social media information from e. g. Facebook). With sophisticated mathematical tools, the adversary can make predictions about the target genome.
Genealogical imputation of multiple relative: this is the more advanced version where the intruders have access to several relatives (say, two grandchildren of the target), which greatly increase the prediction accuracy. This is because the vast majority of genetic commonalities of the two grandchildren comes from the target grandparent.
How individuals can protect their genetic information
Because of the many techniques that adversaries can employ to breach genetic privacy and because of the many challenges that exists for the safe storage of genetic information, researchers and technicians have developed a number of tools that can be used to safeguard genetic privacy. These include access control so that not just anyone can access the data, k-anonymity which makes the quasi-identifiers more fuzzy, differential privacy which is based on noise addition, homomorphic encryption which allows accurate analysis on encrypted data, secure multiparty computation that safeguards private information in the context of joint computation and one-sided database searches that disallows the acquisition of genetic information from personal identity.
Access control: the easiest method to ensure adequate privacy of genetic information is to make sure that only authorized personnel can access it. Instead of having it accessible for everyone on the Internet, specialized committees can investigate applicants and only approve those that pass certain criteria.
k-anonymity: although full anonymization by complete removal of all metadata would substantially reduce research ability, it might be enough to reduce the resolution of quasi-identifiers so that a minimum of k-1 samples have exactly the same identifiers. The selection of the value for k is based on a trade-off between privacy (higher k) and data usability (lower k). However, k-anonymity approaches can still be exploited by ADAD, since it only requires the matching of a sample from a known individual to a sample associated with sensitive information, regardless of whatever quasi-identifiers are present for that sample.
Differential privacy: this approach is based on adding noise to the data in such a way that an adversary can never be sure that the database includes the target, since it would look exactly the same whether or not the target could be found in it. The benefit with this approach is that it is safe against adversaries regardless of the level of prior knowledge. However, in some circumstances, the level of noise added might be so large as to make the data considerably less useful for research purposes.
Homomorphic encryption: this is a special kind of encryption that allows analysis of encrypted data and the decryption of the results returns the same information had the analysis been carried out on unencrypted data. This allows the outsourcing of analysis without risking leakage of sensitive information.
Secure multiparty computation: this allows two users to carry out joint analyses on private input data without leaking this private data to the other party. On method that employs this technique is based on sending encrypted sequences to a cloud service that allows sequence alignment based on matches between strings. So instead of “AATGCC” aligning to “AATGCC”, whatever the symbol that this becomes after encryption is matched instead. This puts the vast majority of computational effort, although it takes several times longer.
One-sided database searches: forensic databases might be made more secure if it only allows searches in the “genetic information -> personal identity” and block attempts to go from “personal identity -> genetic information”.



